Roadmap for Implementing Biosecurity and Biodefense Policy in the United States
Overview
The U.S. policy landscape for countering biological threats is split into two main groups: 1) biosecurity, which specifically focuses on preventing theft, diversion, or deliberate malicious use of biological sciences knowledge, skills, materials, and technologies to cause harm; and 2) biodefense, which involves the development of capabilities and knowledge to assess, detect, monitor, respond to, and attribute biological threats. This project involved the first ever systems-based analysis of the entire U.S. biosecurity and biodefense policy landscape, which enabled greater understanding of the functional relationships between policies as of 2017. These analyses, along with reviews of methodologies for measuring policy implementation and historical case studies to better understand factors that lead to opportunity costs, informed the development of a roadmap for implementing biosecurity and biodefense policies that leverages science and technology advances and minimizes security risks. In addition to the roadmap, this study presents two analytic frameworks for evaluating policy implementation and analyzing opportunity costs.
-
- GRYPHON STAFF | Dr. Corey Meyer, Dr. Emily Billings
- FORMER GRYPHON STAFF | Dr. Kavita Berger (Director, Board on Life Sciences at the National Academies of Sciences, Engineering, and Medicine)
- PROJECT PARTNERS | Dr. Diane DiEuliis (National Defense University), Dr. Venkat Rao (Parsons)
- FUNDER | U.S. Air Force Academy and Defense Threat Reduction Agency: Project on Advanced Systems and Concepts for Countering Weapons of Mass Destruction
Resources
Relational map of U.S. biosecurity and biodefense policy by policy subject
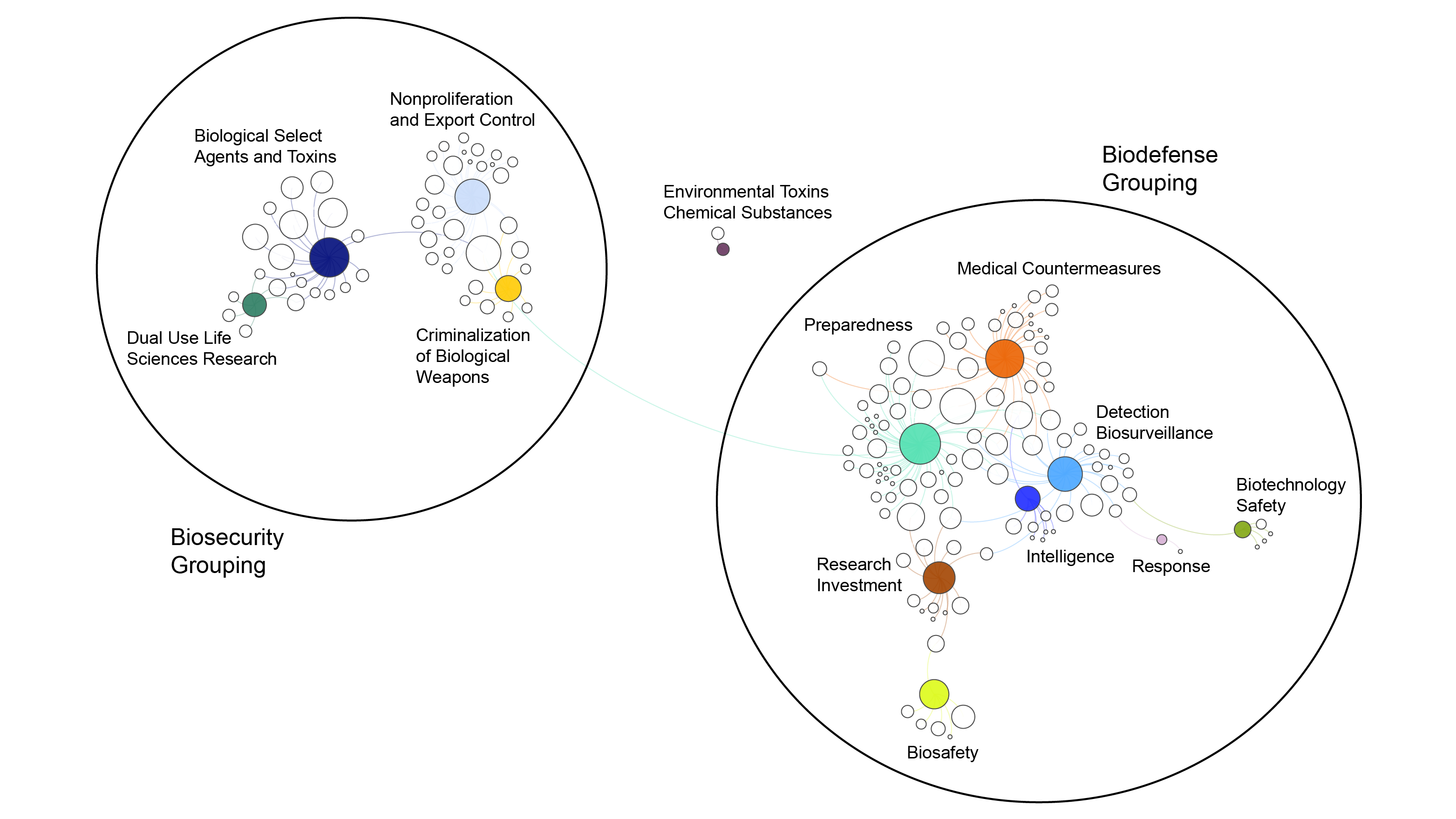
Figure 1. This figure shows a relational map of U.S. biosecurity and biodefense policy by policy subject. Each white circle is a unique U.S. Code, international agreement or partnership, Executive or agency-level policy, program activity (if not already associated with a U.S. Code, international partnership, or agency-level policy), guidance, or guidelines. The colored circles signify subject area. The size of the nodes reflects the number of policies associated with each subject area and the distance between nodes reflects the degree to which policies are linked based on the underlying relational database. The lines reflect direct relationships between policies and subject areas based only on existing policies before 2018. This map does not reflect associations of subject area based on conceptual similarities, but rather associations by direct links between existing policies.
Schematic Map of U.S. Biosecurity and Biodefense Policies to U.S. Biodefense Objectives
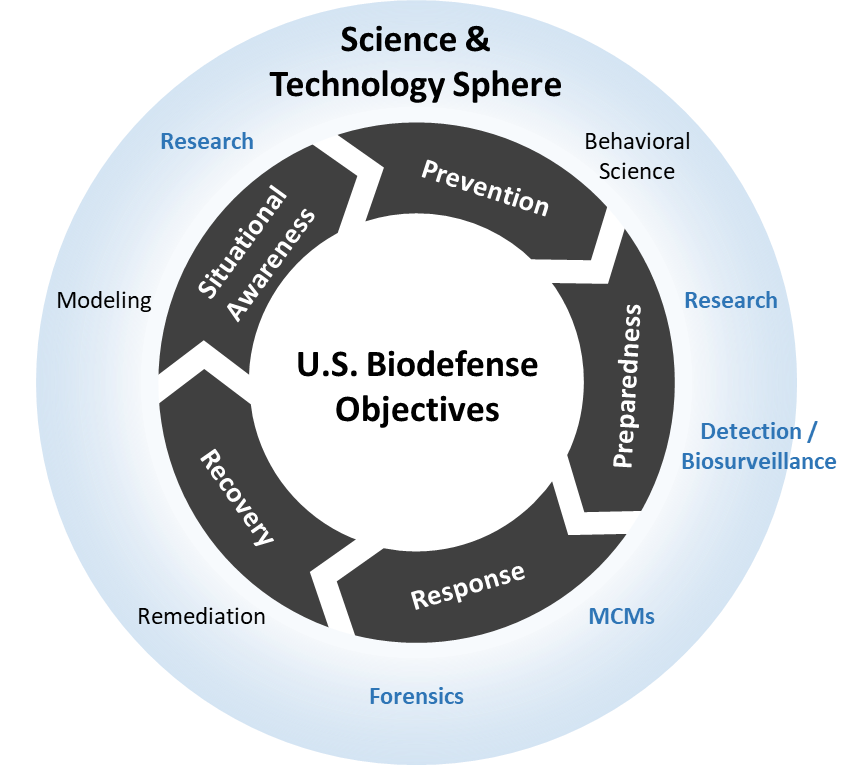
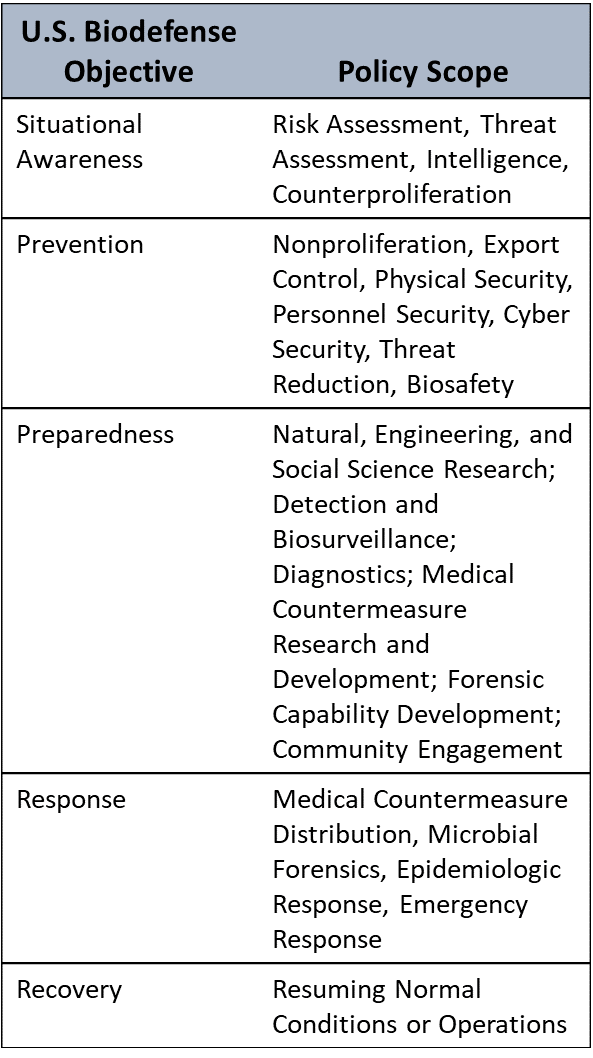
Figure 2. These objectives were derived from combining the core components of Homeland Security Presidential Directive 10/Biodefense for the 21st Century, Presidential Policy Directive 2/National Strategy for Countering Biological Threats, and other relevant policies. The science and technology categories (blue text) were derived from policy documents and funding announcements for research and development for biodefense. Similarly, the policy scope was derived from policy documents and discourse over the past 18 years.
Potential indirect effects of U.S. biosecurity and biodefense policies on U.S. biodefense objectives
Figure 3. This figure shows the potential indirect effects of U.S. biosecurity and biodefense policies on U.S. biodefense objectives. Each circle is a unique U.S. Code, international agreement or partnership, Executive or agency-level policy, program activity (if not already associated with a U.S. Code, international partnership, or agency-level policy), guidance, or guidelines. The white circles represent the direct biodefense objectives (columns) that policies in each of the subject area categories (rows) addresses. The colored circles indicate possible indirect effects of the policies in the subject area categories to biodefense objectives. The green circles indicate capability-building activities. The pink circles indicate requirements, the red circles indicate regulations, and the burgundy circles indicate restrictions, all of which seek to promote biosecurity and biosafety activities. The blue circles are policies that criminalize development and/or use of biological weapons or their delivery systems. Horizontal comparisons between the purpose of a policy (i.e., direct biodefense objective) and the potential indirect effects of a policy should be made.